Here we describe the TRiP platform for the generation of a genome scale collection of Drosophila RNAi stocks. For more details on stocks and plasmids please visit the Reagents page.
- Overview of TRiP stock production (below)
- Knockdown vectors (to understand the strategy or make your own)
- Cloning and sequencing (for confirmation of constructs and fly stocks)
- Using TRiP lines--genetic crosses and strategies
- Using the Gal4-UAS approach for knockdown in the germline
- Using C911 RNAi for testing off target effects
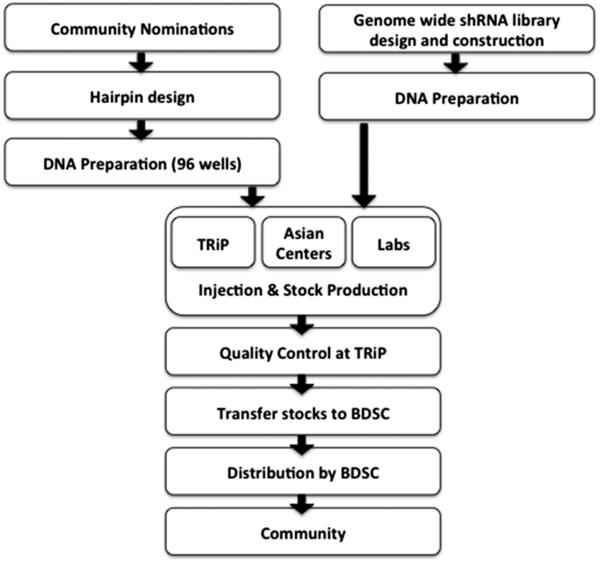
The TRiP production pipeline
Targeted transgenic RNA
TRiP stocks use the Gal4/UAS system (Brand and Perrimon, 1993) to induce the specific expression of a hairpin structure, which silences target gene expression via RNA interference (RNAi).
Hairpin-containing transgenes are inserted via site-specific recombination (Groth et al. 2004) into optimal genomic loci. The main advantage of this method, in addition to its relatively simple design and fast execution time, is that it allows spatial and temporal control of the knockdown construct.
Targeted vs. P-element based integration methods
- The frequency of recovering transformants using the integrase method, either following co-injection of integrase mRNA or by injecting into the nanos-integrase fly strain, is almost five-fold higher than in conventional P-element transformation
- Establishment of the lines is greatly facilitated as no mapping of the transformants to a specific chromosome is needed
- Unlike P-element-based methods, insertions into the attP landing site are homozygous viable
Strategy for selecting Optimal attP Insertion sites
- To avoid the variability associated with some P-element-based approaches, all TRiP vectors are inserted via the phiC31 targeted integration method (Groth et al., 2004) into genomic loci known to be optimal for expression
- To identify attP landing sites that permit optimal transgene regulation and expression, we generated a set of over 20 attP landing sites randomly distributed across the genome
- Used an integrated UAS::luciferase reporter to measure basal and Gal4-mediated expression at these sites (Markstein et al., 2008)
- Determined which of these landing site loci permit high levels of inducible expression by measuring the levels of luciferase activity induced by various ubiquitous or tissue-specific Gal4 drivers
- Based on these analyses, we selected attP40 (located at 25C6 on the second chromosome) and attP2 (located at 68A4 on the third chromosome) as the optimal sites for the integration of hairpin constructs on the second and third chromosomes, respectively
.
Levels of induction at various attP loci
