Protocol for dsRNA Synthesis
We routinely produce dsRNA by in vitro transcription of a PCR generated DNA template containing the T7 promoter sequence on both ends. It is also possible to produce dsRNA using PCR generated DNA templates containing either the T7 & SP6 or the T7 & T3 promoters on either. This method is less efficient, especially when working on a large scale.
**All work should be done in a sterile, RNase free environment, using only sterile, RNase free solutions and materials, and while wearing gloves to reduce contamination.
Template Design
Templates can be generated by PCR on cDNA (including ESTs from BDGP), genomic DNA, or first strand RT-cDNA. Most of the dsRNA should correspond to exons but dsRNA with two or more exons interrupted by introns will also work well. We generally aim for ~300-600 bp products although RNAi with products ranging from 150-3000bp have been shown to work. The target sequence should not contain complete 19-mer homology to other genes or your dsRNA could be non-specific. See Kulkarni, et. al. [references] for further details of the issues surrounding template selection. The DRSC uses genomic DNA as the template. To create your own design, we suggest using the SnapDragon tool for primer design. After choosing the primer sequence, add the T7 promoter sequence TAATACGACTCACTATAGGG to the 5' end of both primers.
PCR Amplification from PCR Templates
*5µL of template PCR are will be shipped to you. We highly recommend that you re-PCR with primers recognizing T7 (see above) to generate more template, before using the PCR template in an IVT reaction. After you've PCR'ed to generate more template, dilute 5µL of template PCR with 10µL of ddH2O. Below are instructions for high-throughput dsRNA synthesis. Single reaction volumes are shown for smaller scale synthesis.
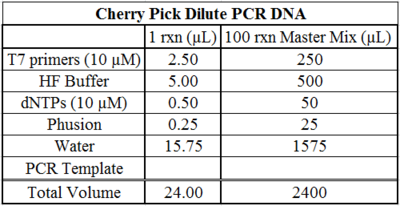
- Produce the above master mix for each 96 well plate you wish to amplify. (The DRSC uses Phusion® Hot Start High-Fidelity DNA Polymerase; NEB M0535).
- Dispense 24 µL of master mix into each well of a 96-well PCR plate.
- Transfer 1 µL of dilute PCR template to its corresponding well.
- Seal the plate using aluminum foil. (At the DRSC we use a Plate-Loc sealer with the 180 adapter for 1.3 seconds). Mix by inverting. Spin down for 1 minute at 2,000 rpm.
-
Run the following PCR program "PCR-57":
- 98°C for 30 seconds
- 98°C for 10 seconds
- 57°C for 20 seconds
- 72°C for 20 seconds
- Repeat steps 2-4 - 35 times
- 72°C for 5 minutes
- 4°C – for ever
- Analyze 5 µL of PCR reaction on a .7% Agarose Gel with the Invitrogen 1kb PLUS ladder.
For most amplicons an annealing temperature of 57°C will give a good band. For bands that appear weak, smeary, negative for a PCR product, or have more than one PCR product per amplicon, a better PCR product can often be produced by either increasing or decreasing the annealing temperature or switching to GC buffer. The DRSC would recommend performing a gradient PCR to find the optimal annealing temperature. If more than one temperature produces a good product, choose the product at the highest temperature.
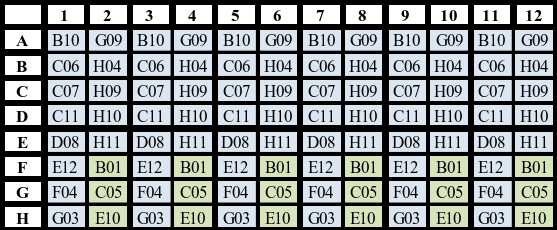
Gradient PCR
- Produce the above master mix for every 16 amplicons you wish to optimize using different annealing temperatures.
- Dispense 144µL of master mix to wells A1-H2.
- Add 6µL dilute PCR template to its corresponding well. Mix thoroughly by pipetting.
- Using a multichannel pipette, transfer 25µL from column 1 [A1-H1] to column 3 [A3-H3]. Repeat transfer from column 1 to columns 5, 7, 9, 11.
- Using a multichannel pipette transfer 25µL from column 2 [A2-H2] to column 4 [A4-H4]. Repeat transfer from column 2 to columns 6, 8, 10, 12.
- Seal the plate using the Plate-Loc or aluminum foil. Mix by inverting. Spin down for 1 minute at 2,000rpm.
-
Run the following PCR program "PCR50-65":
- 98°C for 30 seconds
- 98°C for 10 seconds
- 50—65°C for 20 seconds
- 72°C for 20 seconds
- Repeat steps 2-4 35 times
- 72°C for 5 minutes
- 4°C – for ever
- Analyze 5µL of PCR reaction on a .7% Agarose Gel with 1kb PLUS ladder.
- Consolidate the best amplicons onto one PCR plate. The DRSC recommends that if a better annealing temperature is found; replace the amplicon on your original 96 well "PCR57" plate with the better amplicon. (we make a list of amplicons to replace, vacuum out the bad amplicons and replace them with the higher quality PCR product form the PCR gradient plate)
dsRNA synthesis (IVT reaction)
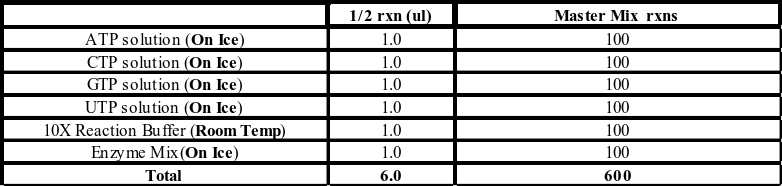
- Produce the above master mix for each 96 well plate you wish to do an IVT reaction on. (We use the Ambion 5X T7 MEGASCRIPT Kit; AMB 1334-5). IVT reaction buffer is extremely high in salt, so make sure the salt is fully dissolved before using the buffer. ½ IVT reactions produce around 30-60µg of dsRNA, enough to produce about 60 384-well assay plates.
- Dispense 6µL of master mix into each well of a 96-well PCR plate.
- Transfer 4µL of amplified PCR product to its corresponding well.
- Seal the plate using the Plate-Loc or aluminum foil. Mix by inverting. Spin down for 1 minute at 2,000rpm.
- Incubate the IVT reaction overnight (~16 hours) at 37°C
- Add 1µL Turbo DNAse [comes in ambion kit]. (The DRSC recommends making a 1:5 dilution and adding 2.5µL to each well using a multichannel pipette)
- Incubate at 37°C for 45 minutes.
-
If using 96 well format go to 8.1. If making smaller volumes go to 8.2. and follow kit instructions for dsRNA cleanup.
- Add water to each well to make a total well volume of 175µL
- If you are doing single reactions, the RNAeasy columns from QIAGEN work very well for IVT clean up. Follow kit instruction and elute in desired volume. Skip to instruction Step 14.
- Transfer samples to Millipore Filter plate (MSNU 03050).
- Vacuum filter on a Millipore vacuum manifold for 45 minutes at 10-15 PSI. [NOTE: PSI recommendation updated from 8-10 to 10-15]
- When all wells are totally dry, add 75µL (or other elution volume) of ddH2O to wells. NOTE: 75µL elution volume will give you a concentration range from 300-800ng/µL. Adjust as needed for your applications.
- Seal in the Plate-Loc or with aluminum foil and shake for 45 minutes.
- Transfer dsRNA from Millipore Filer plate into either a fresh 96well PCR plate or the 96 well plates you prepared the IVT reactions in.
- Run .5µL of dsRNA on a .7% agarose gel and determine quality
- Check dsRNA concentration using a spectrophotometer and dilute accordingly.
